Hi, I’m Kyriakos and my passion is turning data into value.
I bridge the gap between software engineering and drug discovery, translating complex cheminformatics data into practical, data-driven solutions. My experience involves building high-impact tools to tackle complex cheminformatics challenges, like compound selection and reaction prediction. I am a proactive and detail-oriented team player, dedicated to solving scientific problems through collaboration. As a lifelong learner, I am motivated to continuously expand my skills to advance therapeutic development.
I have obtained my Ph.D. degree in Medical Artificial Intelligence from the University of Zurich, affiliated with the University Hospital Zurich and the ETH AI center. I am currently doing a PostDoc at ETH Zurich and Roche in Basel.
Projects in Computational Pharmacology
1. AttentionDDI: Siamese attention-based deep learning method for drug–drug interaction predictions
Improving drug-drug interaction predictions with a novel Transformer-based model.
I developed a novel Deep Learning architecture - AttentionDDI - in PyTorch for the prediction of previously unknown medication side effects. The model is Transformer-based and through an Attention mechanism it provides explainability over which input data modality (chemical structures, side effects, targets, etc.) is weighted more for improved predictions.

2. Drug prescription clusters in the UK Biobank: An assessment of drug-drug interactions and patient outcomes in a large patient cohort
Analysis of patient clusters and their DDIs in a large real-world dataset.
I created patient clusters, according to their drug prescription history, and analyzed the prevalence of DDIs in each cluster, which may influence their five-year mortality. I examined whether specific drug combinations may lead to additional health effects in the UK Biobank participants. For this project I employed the Scikit-Learn, RapidsAI, Scipy and statsmodels data science libraries.

A novel drug-drug-cell line synergy prediction model based on Graph Neural Networks.
A novel Graph Deep Learning architecture - DDoS - in PyTorch for the prediction of synergistic effects of drugs in specific cell lines. The model learns task-specific representations of molecules (drug chemical structures) through a Graph Neural Network and takes advantage of gene expression features of cell lines in order to improve drug combination synergy predictions.

Projects in Bioinformatics
1. Long non-coding RNA structure estimations for cancer genomics
RNA secondary structure calculation for the identification of cancer driver lncRNAs.
Computational methods that have aimed to predict cancer driver lncRNA genes have thus far relied on scores that do not specifically reflect the lncRNA biology. Therefore, Functional Impact scores, such as conservation scores or base pairing probabilties from RNA secondary structure can be included to improve those predictions. In this project I calculated lncRNA base pairing probabilities and correlated them with conservation scores to examine whether secondary structure is suggestive of gene function. Tools used for this project: Bash (awk, bedtools, bwtool, etc.), Python (Snakemake), R, RNAfold/RNAsnp

2. EM algorithm for genomic methylation profiles
Implementation of an Expectation-Maximization algorithm for promoter states in C. elegans.
In C. elegans the expression of X-linked genes is downregulated by half by modifying transcription initiation. Here, an EM algorithm was applied to a dataset containing the positions and the methylation status of Cytosines around the transcription start site (TSS) of various genes. The algorithm is written in R.

Projects in Molecular Biology
1. Effects of tubulin-tyrosine ligase on alpha-tubulin
Four molecular biology experiments for the effects of the TTL gene.
The alpha-tubulin (α-tubulin) protein is a major structural component of microtubules which in turn are one of the three main filaments of the eukaryotic cytoskeleton. Microtubules are associated with important cellular functions, such as mitosis and meiosis, intracellular transport, cell motility, maintenance of the cell shape and others. The tubulin-tyrosine ligase (TTL) protein functions by ligating a tyrosine molecule at the detyrosinated C-terminal of α-tubulin. This process can have an effect on microtubule stability and subsequently on the cellular functions which are depended on the microtubule dynamics. In this work four experiments were performed which try to elucidate the consequences of engineered TTL genes in Drosophila melanogaster. The experiments include:
- Measuring viability and fertility of Drosophila flies
- Confocal microscopy of Drosophila ovaries
- Western blot of tyrosinated α-tubulin
- TTL-GFP fusion gene cloning

Projects in the Cloud - Big Data Analytics
1. From traditional Data Warehouse to modern cloud-based Big Data Analytics
Transitioning to a modern and scalable cloud solution to enable Big Data Analytics.
I empowered my team to transition from a in-house DWH solution to a scalable cloud solution, Azure Databricks. This enabled us to develop modern ETL piplines, and scalable Machine Learning applications.

Projects in Web Development
1. Professional company websites
Professional development of major Swiss company websites.
While working as a software / web developer at fastforward websolutions I was involved in the implementation and maintenance of professional company websites. Some notable examples include:
My responsibilities included web development in HTML/CSS/JS, Magnolia CMS configuration, extension of CMS functionality in Java, setting up REST/SOAP services and planning Agile/Scrum tasks and sprints.

2. Flurnamen Puzzle
Web-based puzzle game for primary school students.
An interactive and educational game where local map regions act as puzzle pieces. The primary school students of Läufelfingen can use it to better learn their region in a fun way by piecing together these puzzle pieces. The game was developed with Processing.js.

Projects in Computer Vision & Computer Graphics
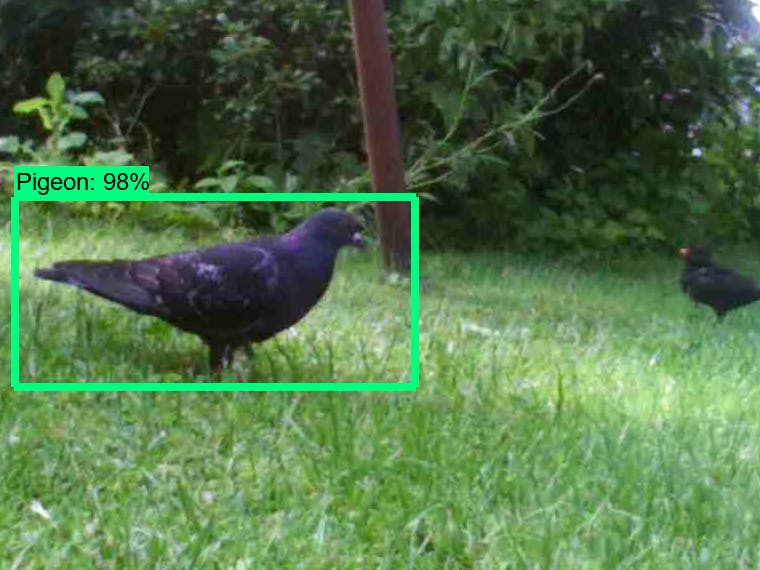
1. Pidgeon Detection
Video analysis algorithm for the detection and positioning of pidgeons.
In this project a video analysis algorithm was developed that utilizes basic computer vision principles. Based on pidgeon characteristics (shape, size, movement, etc.) their position is determined within a video input stream. Written in Python/OpenCV/QT. The video input can be opened in a QT window and areas of interest (detection) can be selected (by drawing polygons). Within these areas the positions of detected pidgeons are reported.
2. L-Systems
Computer generated trees and fractals through L-Systems.
L-Systems are simple rule-based text generation systems. After the text is generated it can be utilized as instructions of Turtle graphics. These turtle graphics then were implemented in Java and OpenGL in order to draw trees and other fractals.